Project
Molecular Dynamics study of mutant IDH1 for in silico drug design against neurological diseases
Recent research has linked the occurrence of mutations on the Isocitrate Dehydrogenase enzyme (IDH1)
to some cancers like low grade glioma, glioblastoma and acute myeloid leukaemia (AML). This has been found to be due to the production of the molecules 2-hydroxyglutarate (2-HG) in abnormal quantities by the mutant IDH1 (mut-IDH1).
Project Details
Project term
November 1, 2020–January 31, 2021
Affiliations
Forschungszentrum Jülich
Institute
Computational Biomedicine Institute
Principal Investigator
Methods
Wild-type IDH1 (wt-IDH1) catalyzes the normal reaction converting isocitrate (ICT) to a-ketoglutarate (aKG) as part of the TCA cycle in glucose metabolism. They are predominantly found in mitochondria and cytoplasm. Mutations on certain amino acids residues of this protein cause it to lose this ability, but results in a ‘new gain of function’ allowing it to catalyze the conversion of aKG to 2-HG. This neomorphic reaction leads to the buildup of abnormal levels of 2-HG in the body, and has been linked with tumorigenesis leading to glioma and AML. Thus, mutant IDH1 (mut-IDH1) could be a potential drug target to treat these diseases. Indeed, many pharmaceutical companies have potential drug candidates in clinical trial stage of development. Although inhibitors have been identified, there is disagreement over the exact catalytic mechanism of the neomorphic reaction. This has impeded a more scientific design of drug candidates. Thus, our aim is to elucidate the mechanism of the neomorphic reaction catalyzed by mut-IDH1 through computational methods in order to aid future drug design efforts.
We planned to do this in two steps, 1) by applying classical Molecular Dynamics (MD) simulations to study the ligand-protein interaction in the wild-type and mutant; and 2) based on frames extracted from the MD simulations, exploit our newly developed MiMiC QM/MM interface to study the transition state and shed light on mechanism of the neomorphic reaction. This project covered the first step, i.e., using MD to study the Michaelis complex of the enzyme-ligand complex as a precursor to the transition state.
Results
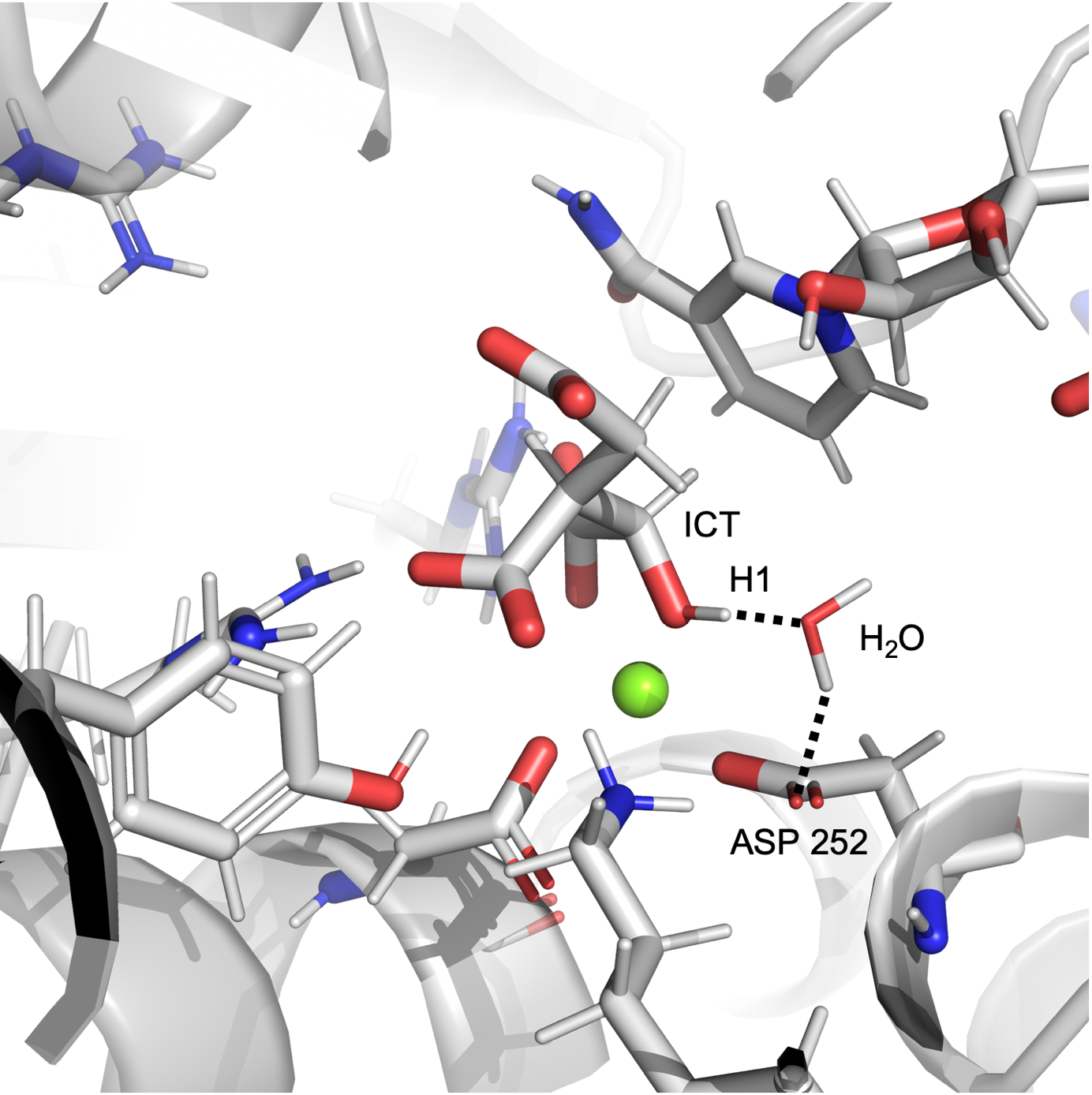
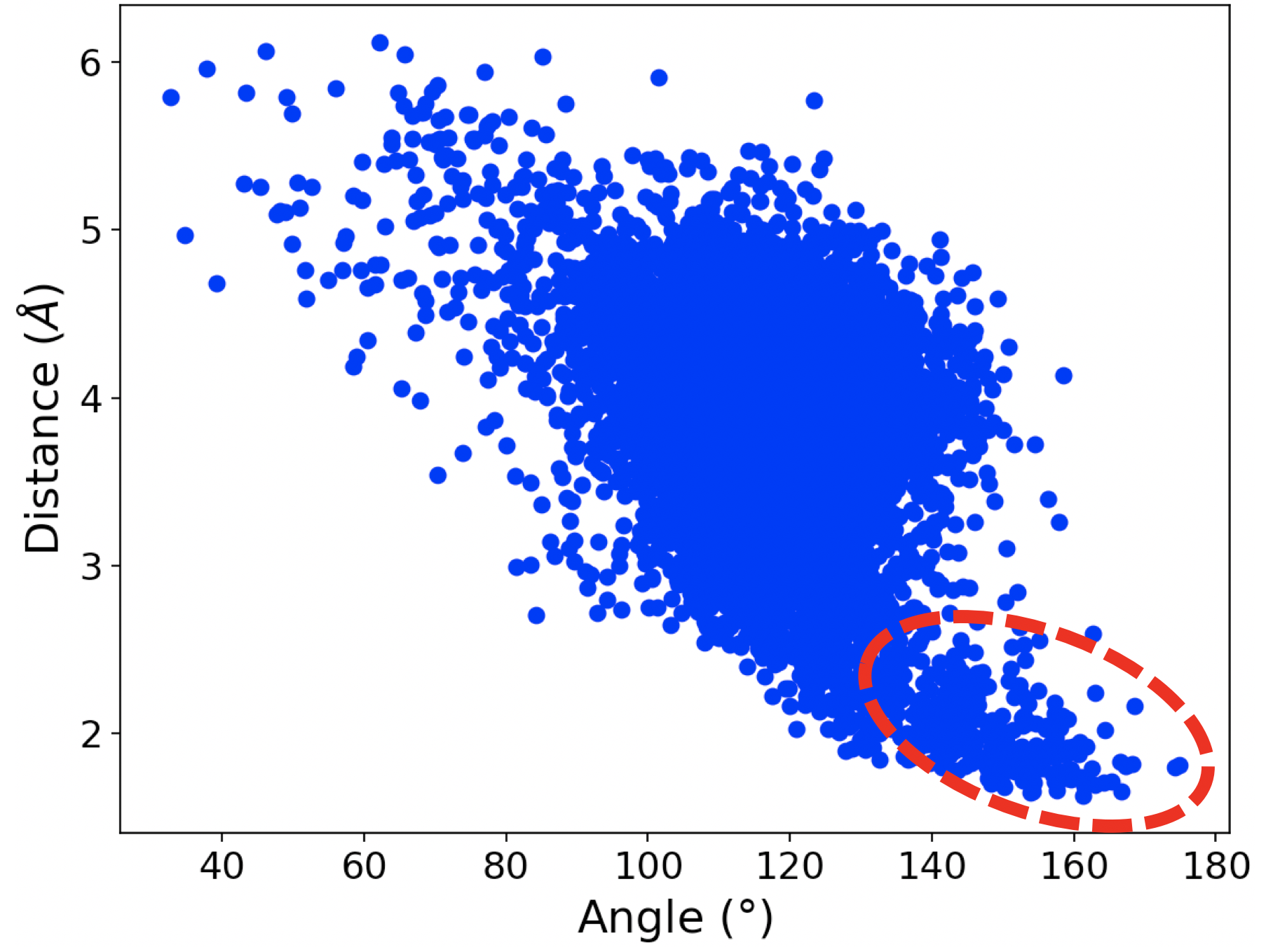
In the simulation of wt-IDH1, we observed a water-aspartate pair interacting with the a-alcoholic hydrogen of ICT as shown in Figure 1a. The plot in Figure 1b shows that at many points in the simulation, the distance and angle of interaction between the alcohol of ICT and the water molecule bound to the aspartate residue Asp 252 exists in the region consistent with hydrogen bonding. This is important because the initiating step of the catalysis is the abstraction of proton from the alcohol of ICT by a base. The hydrogen bond observed between water and the a-alcohol in our simulations suggests that the water-Asp 252 pair could potentially act as the base.
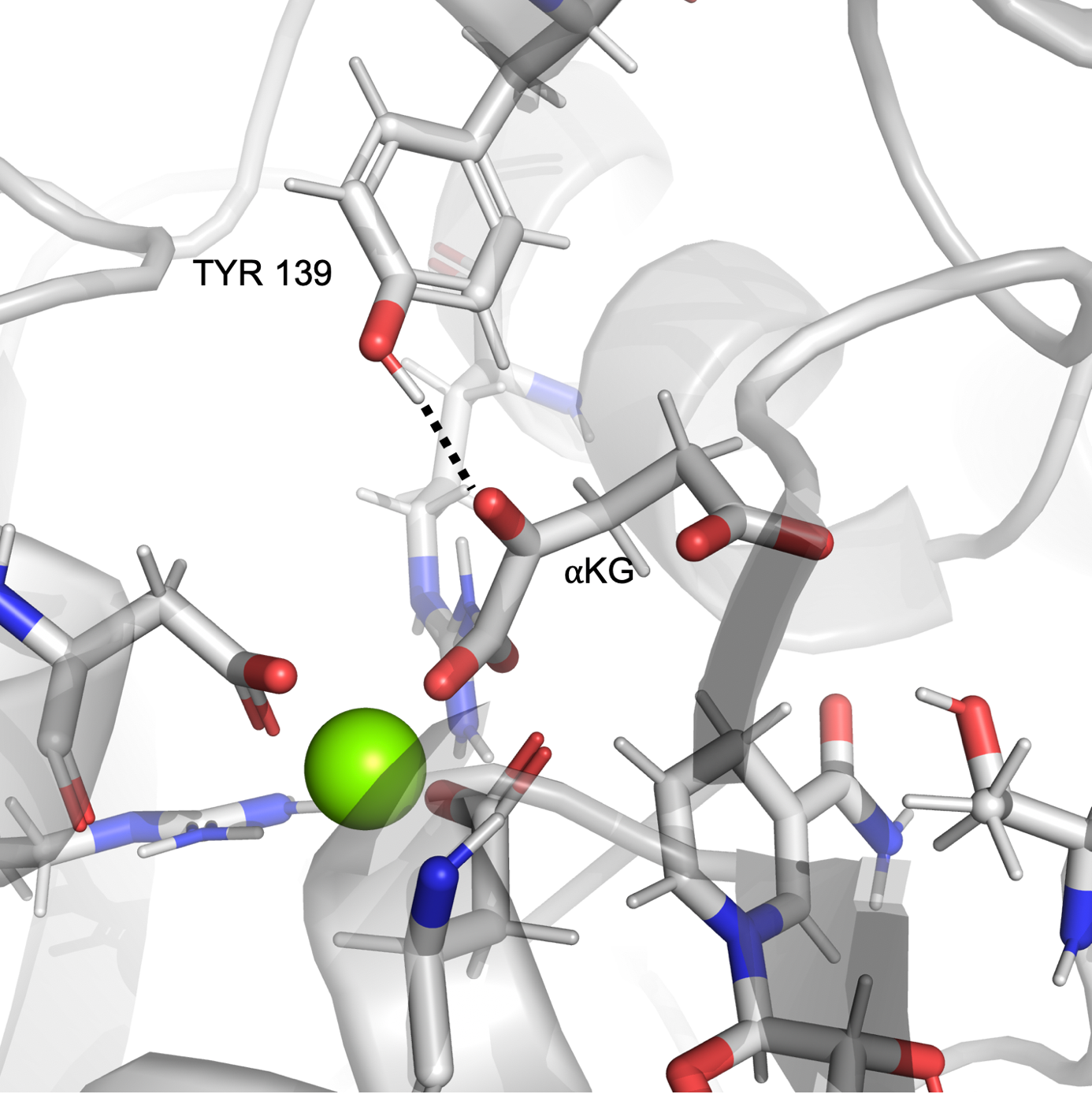
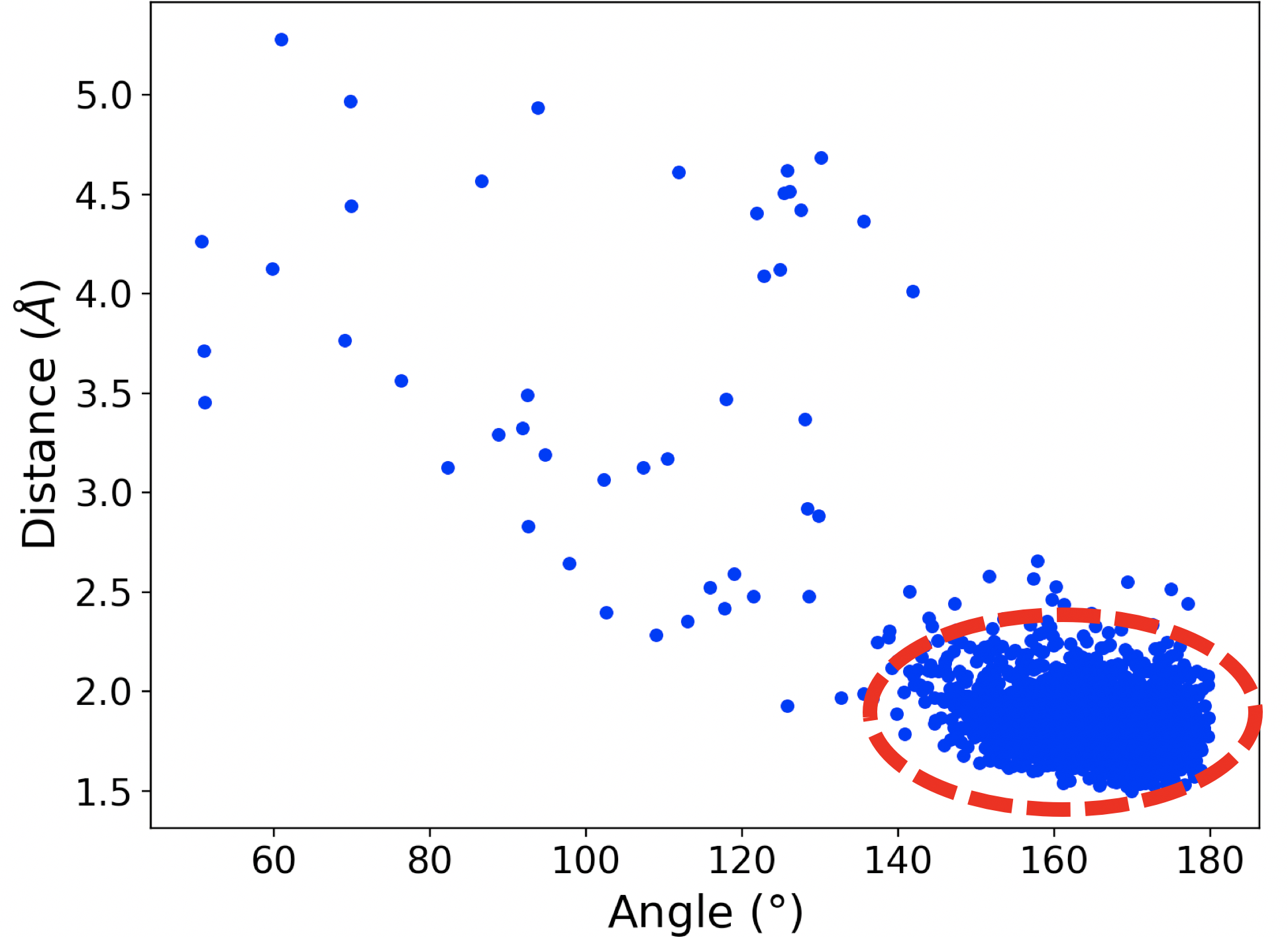
In the case of mut-IDH1, we observed the tyrosine residue Tyr 139 closely interacting with the a-ketone group of aKG as shown in Figure 2a. Similar to Figure 1b, the plot in Figure 2b shows that at many points the simulation exists in the region where the distance and angle of interaction between Tyr 139 and the ketone group of aKG are consistent with hydrogen bonding. In a mirror image of the wt-IDH1 catalysis, the initiating step of the mut-IDH1 catalysis is the protonation of the carbon and oxygen atoms involved in the a-ketone bond of aKG. An acid is required to donate a proton to the oxygen atom. From the interaction observed in the MD simulations, Tyr 139 could potentially act as the acid to protonate the ketonic oxygen and drive the reaction forward.
Discussion
These interesting results on the Michaelis complex and potential acid/base candidates in the IDH1 catalysis will be investigated further in our upcoming QM/MM studies.
Additional Project Information
DFG classification: 201-02 Biophysics, 310 Statistical Physics, Soft Matter, Biological Physics, Nonlinear Dynamics